The chloroplast plays an indispensable role in plant growth and development, but its proteome remains to be further improved. The size and information about Chloroplast Proteome in Arabidopsis varies among different data sources and the number of confirmed chloroplast proteins is obviously inadequate with the comparison of previously prediction. Here a great effort has been made to generate a most comprehensive chloroplast proteome in Arabidopsis and provide new insight into the function of chloroplast.
The structure of chloroplast
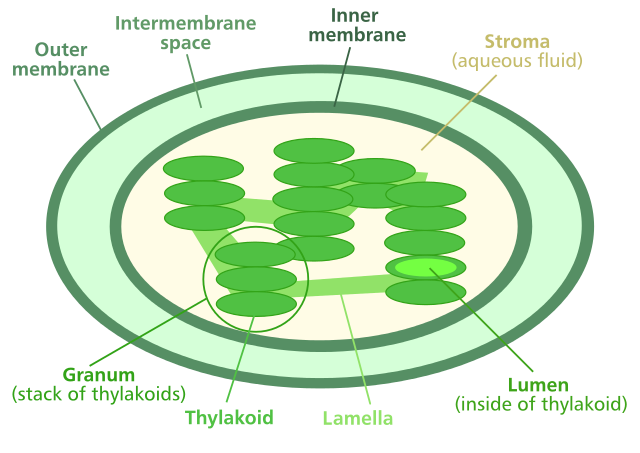
Enlarge the picture
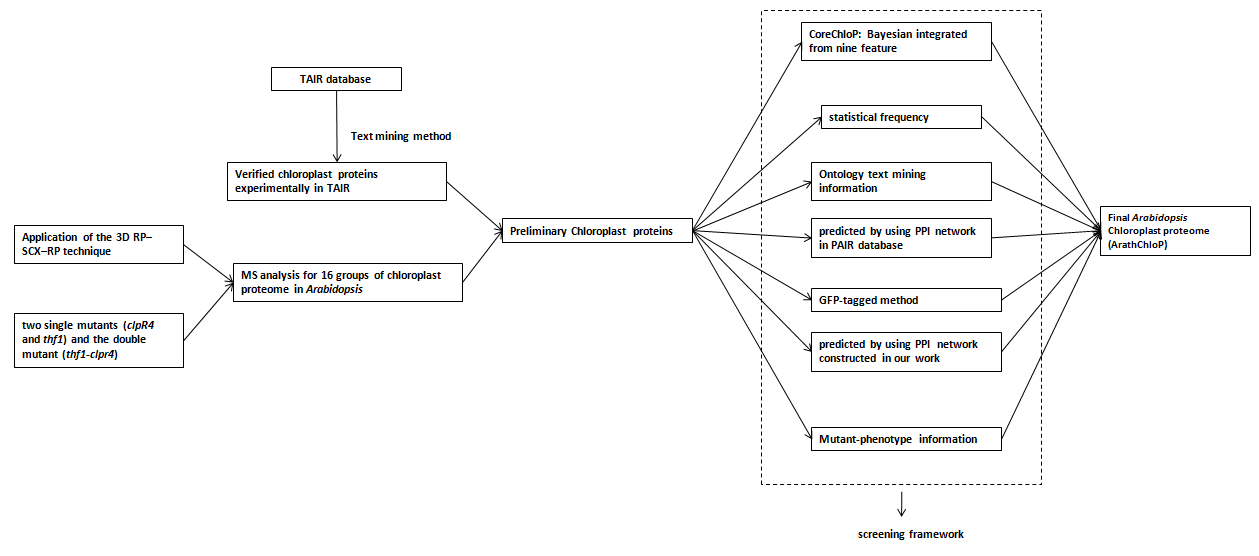

Click the picture for enlarge the picture
In our study, two strategies were constructed to explore the chloroplast proteome. Firstly we employed a computational framework to predict Arabidopsis chloroplast proteins with Naive Bayesian integration model by combining multi-level data generated from bioinformatics tools, ancestry transfer, orthologous mappings and gene ontology annotation, and obtained a set of 2,716 integrated core chloroplast proteins (CoreChloP). Then, we used three Arabidopsis mutants (two single mutants (clpR4 and thf1) and the double mutant (thf1-clpr4)) with enriched low-abundant chloroplast proteins due to the reduced level of FtsHs protease to identify more chloroplast proteins that previous researches were unable to observe and identified 1,948 chloroplast proteins by using mass spectrometry. Finally, 3,134 credible chloroplast proteins (named ArathChloP) were determined by combining the screened MS data and previously reported ones, 349 of them are not reported experimentally in TAIR but validated with multi-approaches in our work including the mutants with clear defects in photosynthesis, GO text mining analysis, PAIR prediction, PPI network evidence, statistical frequency analysis and transgenic plants.
Furthermore, in order to explore the functional proteomics in chloroplast, we adopted a network-based approach to infer functions for those unknown proteins by searching PIN structure and annotated novel functions for 33.42% of the unknown-functional proteins in ArathChloP. We assigned protein functions according to the enriched function term within its level-1 and level-2 neighboring proteins referring to the method described in BioPlex Network..
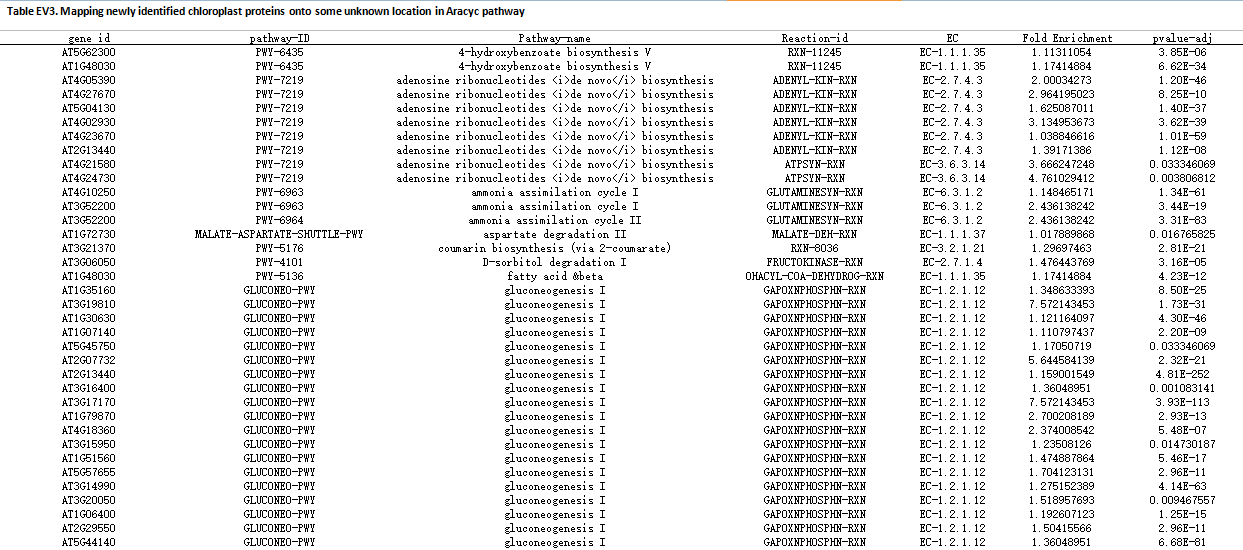
Enlarge the picture
Meanwhile, we mapped 37 newly identified proteins functioning in 52 pathways sites in AraCyc pathway system based on the PPI network according to the enriched pathway at the level-1 and level-2 neighboring proteins.
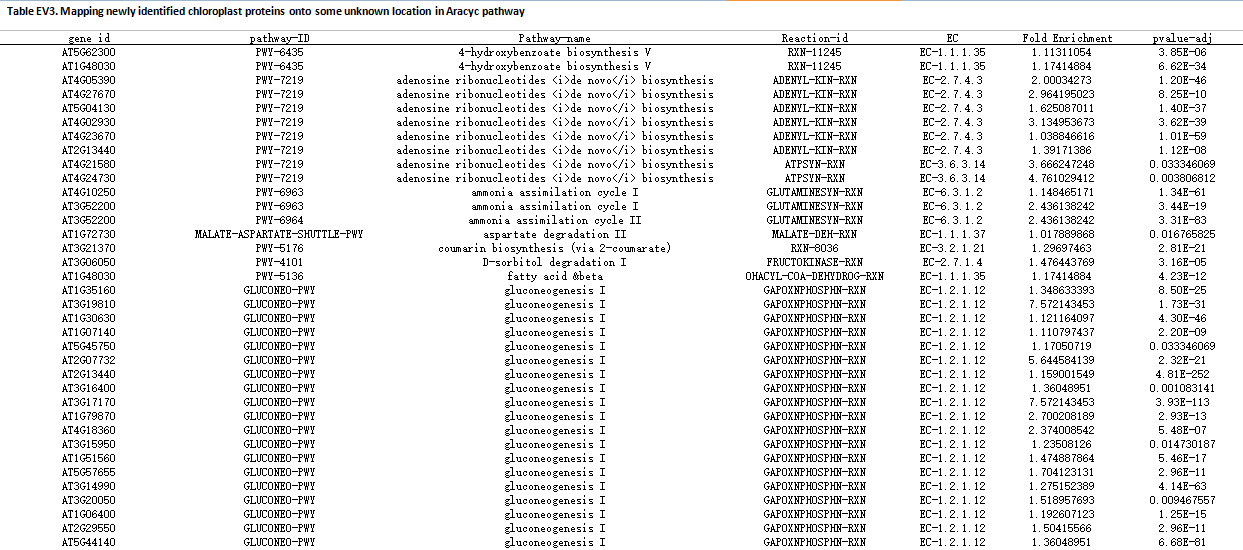
Enlarge the picture
The chloroplast proteome data is available and free access in our AtPID database, you can get the data from the "download" module.