Welcome to DNMIVD
DNA methylation is strongly related to cancer, currently, no resource integratively provides the DNA methylation based diagnostic and prognostic models, e-mQTL (expression–methylation quantitative trait loci), pathway-meQTL (pathway activity-methylation quantitative trait loci), differentially variable and differentially methylated CpGs, survival analysis, as well as functional epigenetic modules for different cancers, which are valuable information for researchers to explore DNA methylation profile from different aspects in cancer.
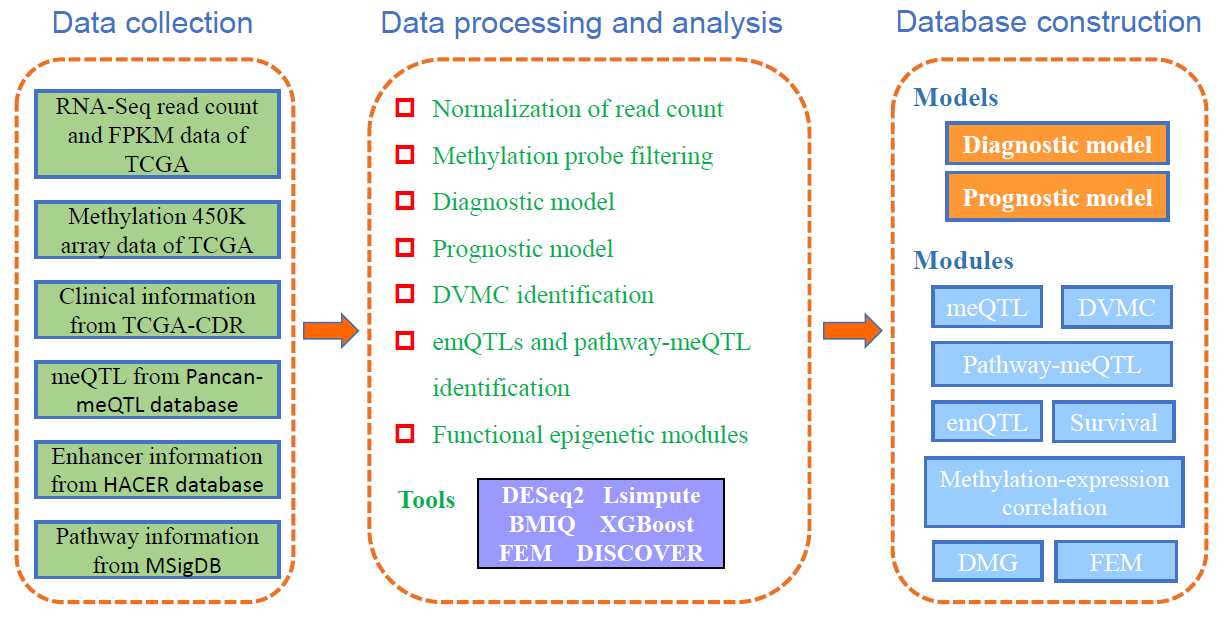
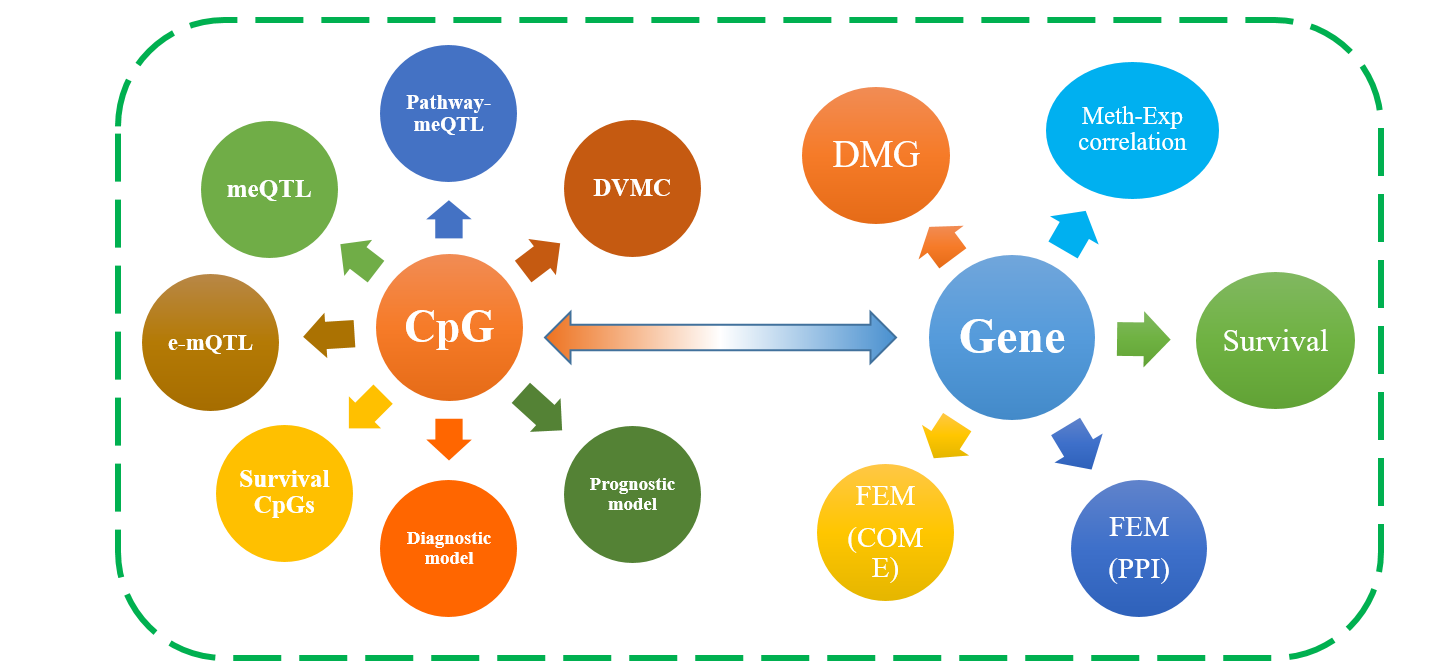
DNMIVD is a comprehensive annotation and interactive visualization database for DNA methylation profile of diverse human cancer constructed with high throughput microarray data from TCGA and GEO databases, and it also integrates some data from Pancan-meQTL and HACER databases. Overall, DNMIVD mainly contains the following important resources:
-
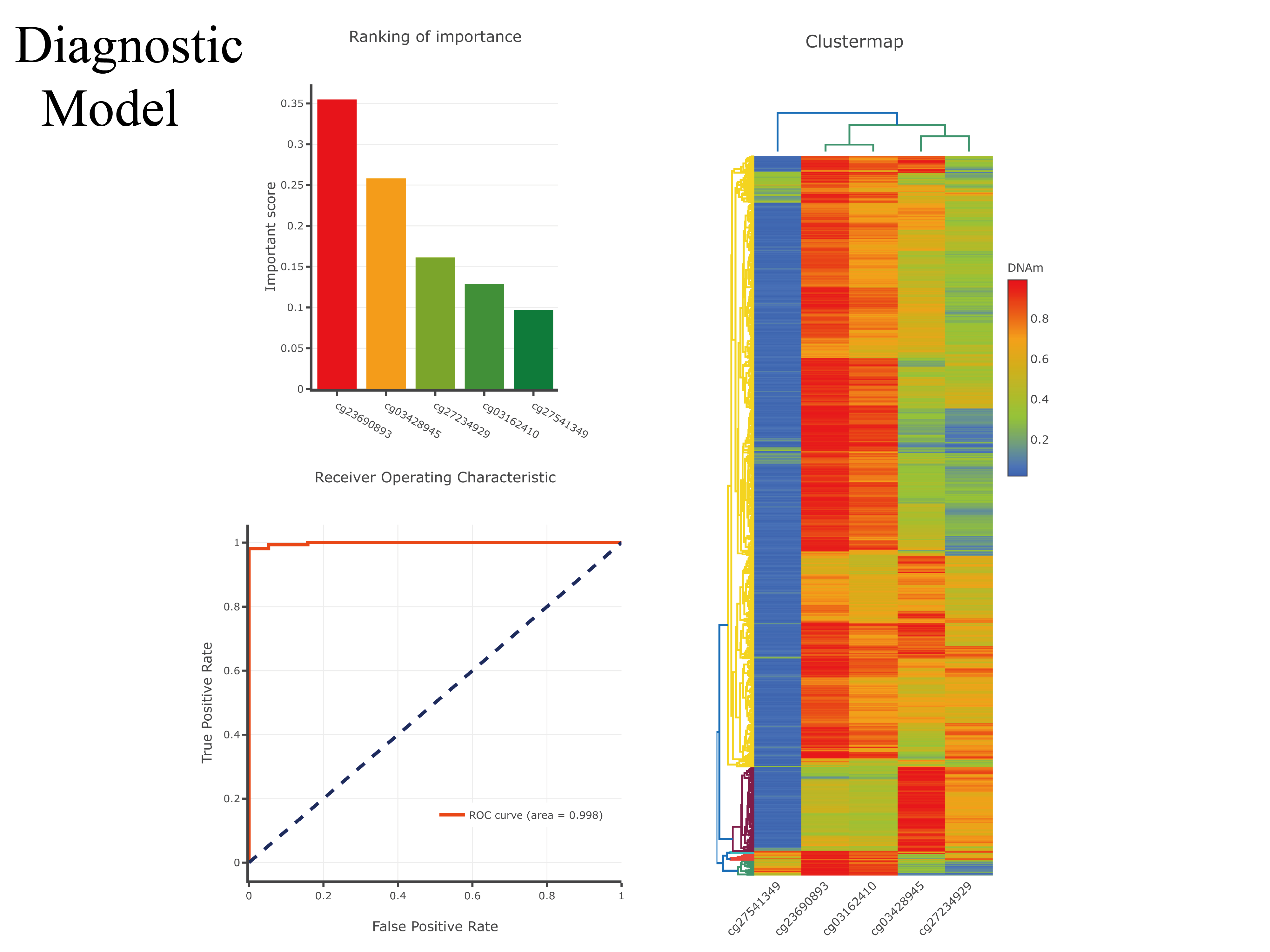
1) diagnostic and prognostic model for 26 The Cancer Genome Atlas (TCGA) cancer types and visualization based on DNA methylation;
-
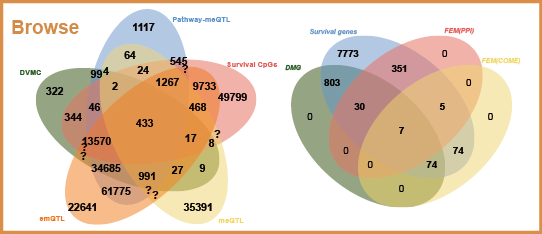
2) meQTL, e-mQTL and pathway-meQTL for diverse cancers;
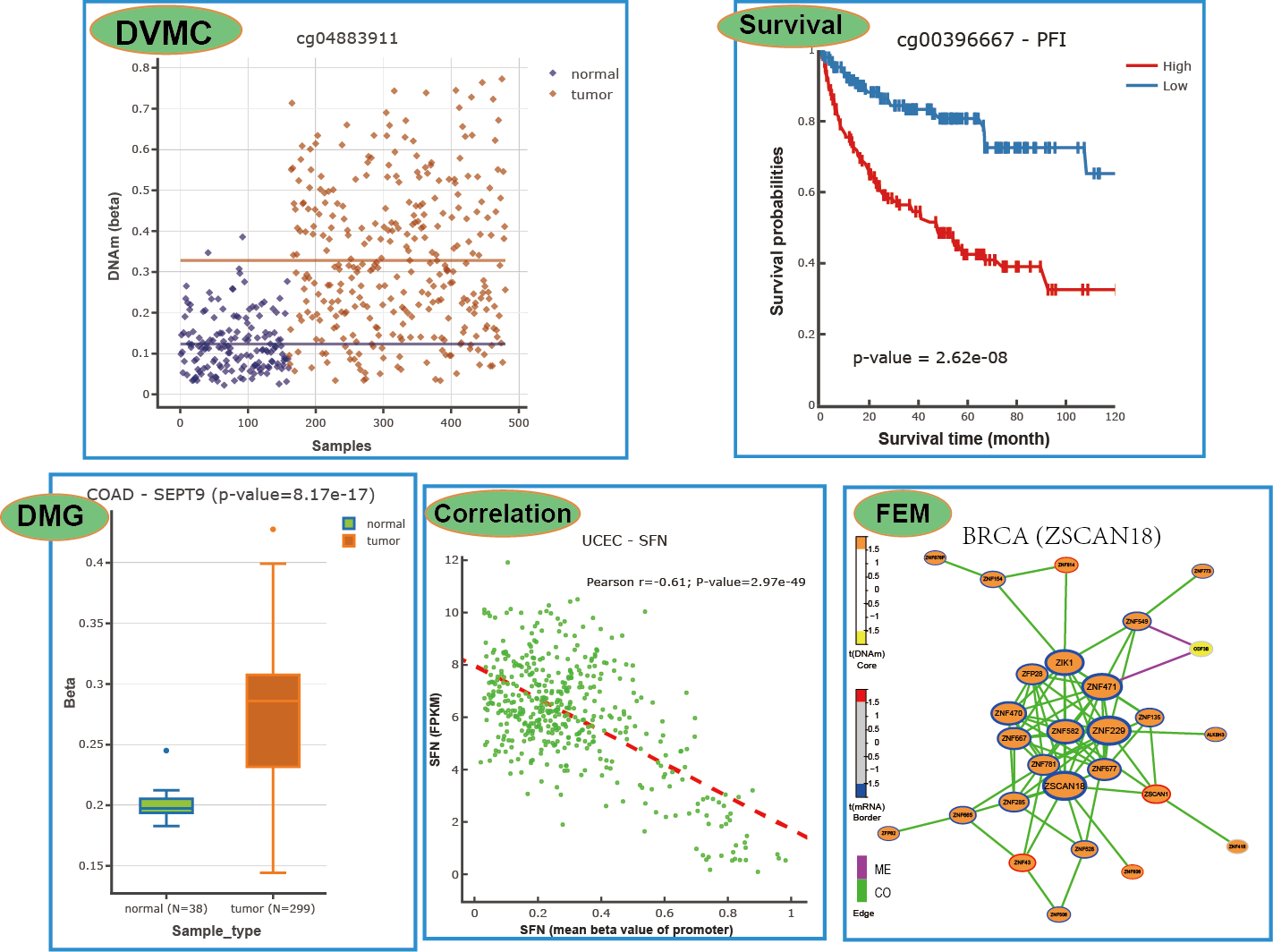
3) Functional Epigenetic Modules (FEM) inferenced by integrating DNA methylation and gene expression data of TCGA cancers, we provide FEM constructed from Protein-Protein Interactions (PPI) and Co-Occurrence and Mutual Exclusive (COME) network for choice;
4) differentially variable and differentially methylated CpGs (DVMCs), differentially methylated genes (DMGs) as well as enhancers information;
5) correlations between methylation level of gene promoter and corresponding gene expression;
6) survival associated CpGs and genes with different endpoints (such as overall survival time, disease-free interval and progression-free interval).
DNMIVD provides a user-friendly interface to browse, search, building model and download data of interest. We believe that this database would become a valuable resource for DNA methylation research in cancers.

Quick Search
Website Compatibility: Google Chrome is recommended
Citation: [2] Wubin Ding, Jiwei Chen, Guoshuang Feng, Geng Chen, Jun Wu, Yongli Guo, Xin Ni, Tieliu Shi, DNMIVD: DNA methylation interactive visualization database, Nucleic Acids Research, 48(D1), D856-D862.[ Full Text ]
Citation: [3] Ding, W., Feng, G., Hu, Y., Chen, G., & Shi, T. (2020). Co-occurrence and mutual exclusivity analysis of DNA methylation reveals distinct subtypes in multiple cancers. Frontiers in cell and developmental biology, 8, 20.[ Full Text ]