1. Introduction
Here, we developed a homology independent ARG identification method (named PLM-ARG) based on an integrated amino acid language model, which enables novel ARG identification and multi-resistance categories annotation.2. Database construction
We incorporated and standardized the collected AGRs from five databases including CARD (Release Date: 05/27/2022) [1], ResFinder (Release Date: 24/05/2021)[2], MEGARes (Release Date: 14/10/2019)[3,4], ARGMiner (Release Version: v1.1.1.A)[5], AMRFinderPlus (Release Date: 11/08/2021)[6] and HMD-ARG-DB[7]. First, considering that various genomic technologies detected the ARGs in these databases, we converted all ARGs with DNA sequence format into UniPort FASTA protein sequence format using EMBOSS tool Transeq. Then, we removed the duplicated ARGs by clustering all their sequences with CD-HIT and discarding duplicates with 100% identity and the same length. Finally, resistance categories of ARGs were assigned based on their conferred antibiotic drug classes with manual correction based on the WHO access, watch, reserve, classification of antibiotics for evaluation and monitoring of use (AWaRe) classification system (https://apps.who.int/iris/rest/bitstreams/1374989/retrieve).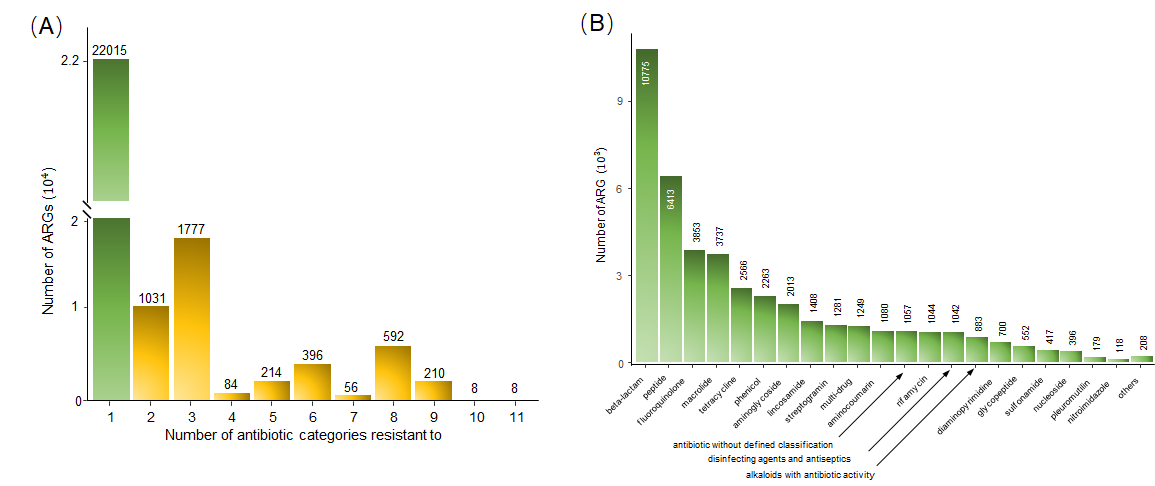 The composition of PLM-ARG database: (A) the relationship between the number of ARGs and number of their antibiotic resistant categories. (B) distribution of ARGs across different resistance categories.
The composition of PLM-ARG database: (A) the relationship between the number of ARGs and number of their antibiotic resistant categories. (B) distribution of ARGs across different resistance categories.
3. Search in PLM-ARG
Input File Format
The input file should follow the standard fasta format. Here is an example: Sample File.
Reference:
[1] Alcock, B.P., Raphenya, A.R., Lau, T.T.Y., Tsang, K.K., Bouchard, M., Edalatmand, A., Huynh, W., Nguyen, A.V., Cheng, A.A., Liu, S. et al. (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res, 48, D517-D525.
[2] Kleinheinz, K.A., Joensen, K.G. and Larsen, M.V. (2014) Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage, 4, e27943.
[3] Doster, E., Lakin, S.M., Dean, C.J., Wolfe, C., Young, J.G., Boucher, C., Belk, K.E., Noyes, N.R. and Morley, P.S. (2020) MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res, 48, D561-D569
[4] Lakin, S.M., Dean, C., Noyes, N.R., Dettenwanger, A., Ross, A.S., Doster, E., Rovira, P., Abdo, Z., Jones, K.L., Ruiz, J. et al. (2017) MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Research, 45, D574-D580.
[5] Arango-Argoty, G.A., Guron, G.K.P., Garner, E., Riquelme, M.V., Heath, L.S., Pruden, A., Vikesland, P.J. and Zhang, L. (2020) ARGminer: a web platform for the crowdsourcing-based curation of antibiotic resistance genes. Bioinformatics, 36, 2966-2973.
[6] Feldgarden, M., Brover, V., Haft, D.H., Prasad, A.B., Slotta, D.J., Tolstoy, I., Tyson, G.H., Zhao, S., Hsu, C.-H., McDermott, P.F.J.A.a. et al. (2019) Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. 63, e00483-00419.
[7] Li, Y., Xu, Z.L., Han, W.K., Cao, H.L., Umarov, R., Yan, A.X., Fan, M., Chen, H., Duarte, C.M., Li, L.H. et al. (2021) HMD-ARG: hierarchical multi-task deep learning for annotating antibiotic resistance genes. Microbiome, 9.
